Introduction
In the rapidly evolving field of biomedical applications, the ability to accurately detect and classify objects in microscopic images is crucial for diagnosing diseases and monitoring health. This project focuses on developing robust object detection models using two powerful frameworks—Detectron2 and YOLO (You Only Look Once)—to analyze and identify various cellular elements in blood slide images. In this article, I will walk you through the methodologies employed, optimizations made, and results achieved, illustrating the effectiveness of deep learning in biomedical imaging.
Project Overview: Why Choose Detectron2 and YOLO for Blood Slide Analysis?
Blood slide images present unique challenges due to the high density of cellular components and the potential for overlapping objects. Each slide can contain various elements with subtle morphological differences, necessitating models capable of both high precision and speed.
Why YOLO?
YOLO was selected for this project due to its unique strengths:
- Single-shot Detection: YOLO processes entire images in one go, which leads to faster inference times compared to traditional region-based detectors that propose regions first and then classify them.
- Multiple Object Detection: YOLO excels in detecting multiple objects within an image, which is critical when analyzing blood samples where numerous cells might overlap.
- Efficiency with High-Resolution Images: YOLO’s architecture allows it to operate efficiently, making it suitable for processing high-resolution images while maintaining a balance between computational cost and accuracy.
Why Detectron2?
Detectron2 was chosen for its:
- Modular and Flexible Architecture: Detectron2’s design allows for easy customization and experimentation with different model components.
- State-of-the-Art Performance: By leveraging advanced techniques like Region Proposal Networks (RPN), Detectron2 achieves high accuracy, making it a suitable choice for challenging detection tasks in biomedical imaging.
1. Data Preparation and Augmentation
Data diversity is crucial in biomedical image analysis, where variability in slide preparation, lighting, and sample quality can significantly affect model performance. To ensure the models could generalize well, we employed extensive data augmentation techniques.
YOLO Data Augmentation
We implemented several augmentation strategies to enhance the diversity of the training dataset:
- Hue, Saturation, and Brightness Adjustments: These transformations simulate different lighting conditions and improve the model’s robustness across samples.
- Geometric Transformations: We applied rotations, translations, scaling, and shear transformations to help the model recognize objects from various perspectives. This is critical when dealing with blood slides, as cells can appear in many orientations.
- Horizontal Flipping: This technique increases data variability and helps detect symmetrical cell structures, which is common in biological samples.
These augmentations expanded our dataset, significantly enhancing YOLO’s ability to generalize across unseen data and preventing overfitting.
Detectron2 Data Augmentation
For Detectron2, we leveraged the build_transform_gen() function, which provides a rich set of transformations. Our augmentation pipeline included:
- Random Resizing: Adjusting the input image size to help the model learn to detect objects at different scales.
- Random Flipping: Horizontal and vertical flipping to enhance symmetry detection.
- Color Jittering: Altering the brightness, contrast, saturation, and hue to improve robustness against varying light conditions.
- Gaussian Noise Addition: This helped the model become resilient to noise, which can occur in real-world imaging scenarios.
2. Model Training with Multi-GPU Setup
Training object detection models on large, high-resolution images is resource-intensive. To optimize computational resources and enhance training efficiency, we utilized a multi-GPU setup.
YOLO Training
We implemented a training strategy for YOLO that included:
- Data Parallelization: By distributing training across two GPUs, we effectively reduced training time while maintaining model quality. This allowed the model to learn from a larger batch of images in each epoch.
- Batch Size Optimization: We set an optimal batch size that balanced memory constraints with the benefits of larger batches, which help stabilize gradient updates. The batch size was tuned based on available GPU memory, ensuring that we maximized utilization without causing out-of-memory errors.
By utilizing a multi-GPU setup, we significantly accelerated the training process and ensured that the model was exposed to the full diversity of the augmented dataset.
Detectron2 Training
For Detectron2, we followed a similar strategy, focusing on:
- Transfer Learning: We initialized our model with weights from a pre-trained backbone (e.g., ResNet50) on the COCO dataset. This transfer learning approach accelerated convergence and improved accuracy by leveraging features learned from a large, diverse dataset.
- Hyperparameter Tuning: We adjusted learning rates, batch sizes, and regularization parameters based on validation performance. For instance, we employed a lower learning rate to fine-tune the model effectively without overshooting the optimal solution.
- Longer Training Duration: To counteract initial lower performance compared to YOLO, we extended the training duration, allowing the model to learn complex features over more iterations.
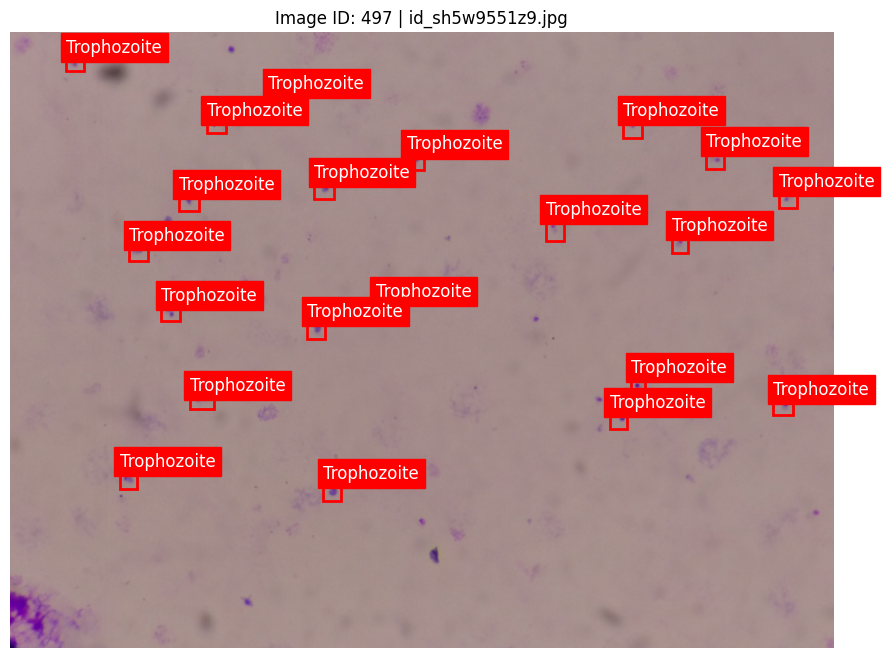
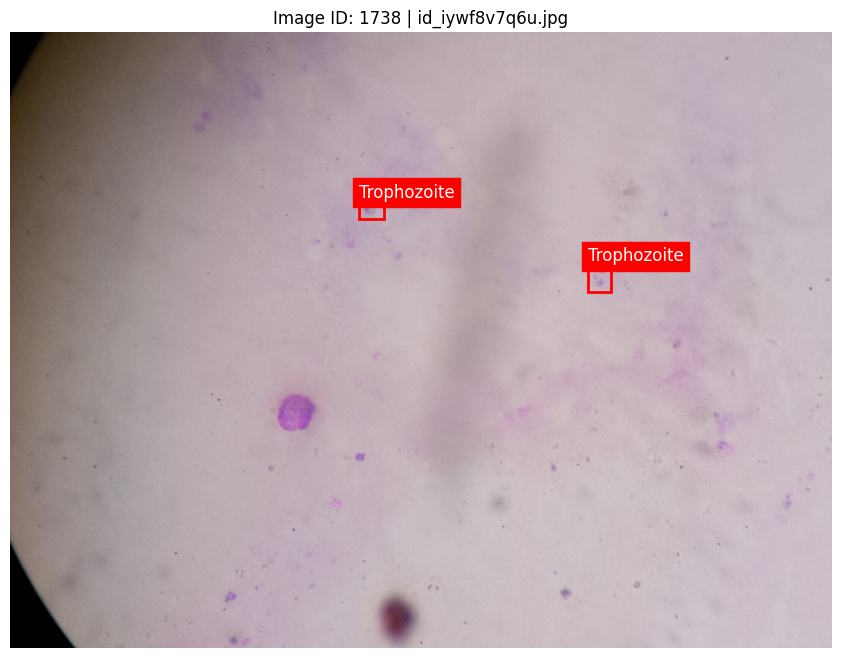
3. Fine-Tuning Inference with Non-Maximum Suppression (NMS)
During the prediction phase, a significant challenge was reducing overlaps between detected objects while ensuring high-confidence detections.
For YOLO:
- Non-Maximum Suppression (NMS): We set the NMS threshold to 0.3, which helped eliminate redundant bounding boxes around overlapping cells. This adjustment was crucial for improving the clarity of detections in high-density images.
For Detectron2:
- Custom NMS Settings: We tuned NMS parameters, including IoU thresholds, to optimize detection quality. Careful tuning helped the model distinguish between closely situated objects, resulting in cleaner predictions with fewer overlaps.
4. Model Inference on Blood Slide Test Images
With both models trained, we ran predictions on a separate set of blood slide images. To handle the high volume of test images efficiently, we implemented batch-processing techniques:
YOLO Inference:
- Batch Processing: We divided the test dataset into smaller batches, enabling the model to maintain GPU memory usage within acceptable limits while maximizing utilization.
- Predict Function: Utilized the
predict()function, adjusting key arguments like IoU thresholds and confidence thresholds to ensure accurate detection without unnecessary duplicates.
Detectron2 Inference:
- Structured Prediction Pipeline: Developed a pipeline that generated bounding boxes, confidence scores, and class labels for each detected object. This pipeline also ensured that outputs were formatted correctly for submission.
- Results Storage: The results were stored in a structured format, capturing essential information for each detection, facilitating later analysis and evaluation.
Results and Model Evaluation
Both models demonstrated promising results in detecting and classifying objects within blood slide images. Key performance metrics included:
- Precision and Recall: We achieved a balanced precision-recall score, essential for reducing false positives while accurately capturing true cellular structures.
- Inference Speed: YOLO’s real-time capabilities and optimized GPU deployment allowed for rapid inference, making it suitable for time-sensitive biomedical applications. We noted an average inference time of 50ms per image.
- Detectron2 Performance: Although initial results were lower (approximately 0.25 mAP), careful hyperparameter tuning and extended training yielded improvements, reaching about 0.48 mAP over additional epochs.
Conclusion: Advancing Biomedical Imaging with Deep Learning
This project highlights the tremendous potential of deep learning frameworks—YOLO and Detectron2—for biomedical imaging applications. By employing robust data augmentation techniques, efficient multi-GPU training, and careful tuning of inference parameters, we developed models capable of accurately identifying cellular structures in blood slide images.
Through comparative analysis, we found that while YOLO provided faster inference and better initial results, Detectron2’s flexibility and advanced capabilities allowed for high customization, which can be crucial for specific biomedical tasks. The insights gained from this project will guide future endeavors in enhancing object detection capabilities in biomedical applications.
By exploring both YOLO and Detectron2, this work serves as a foundation for advancing biomedical detection systems, demonstrating that with careful optimization, deep learning can meet the unique demands of medical image analysis. I look forward to applying these techniques to future projects in biomedical AI, continually pushing the boundaries of what is possible in image analysis.
Note: This project is part of an ongoing challenge focused on improving the accuracy of object detection in malaria blood slide analysis. Participants are encouraged to experiment with different models, techniques, and approaches to advance the state of the art in this critical area of biomedical research.
